HYPHY stuff: all files need for this tutorial are here
Identification of pervasive positive selection in cone snail venom
1. Get the file Conus.01.fas * This is our translated alignment

2. We will now do the FUBAR analysis * This is a site specific method for detecting selection at the amino acid level, helpful in arms race scenarios
3. Go to http://www.datamonkey.org/dataupload.php
**4. ** Select choose file, select Conus.01.fas * Then, press Upload
5. If your data is no bueno, this is where it will let you know * It accepts fasta alignments and nexus files * Also listed here should be information about the alignments * If all is well, press Proceed to the analysis menu
6. Under Method select FUBAR
7. For now leave everything else alone and just press Run * If you had a nexus file, you could specify your own tree in Newick format * For now we’ll just let it use a generated neighbor joining tree 😢
8. The results will look like the following 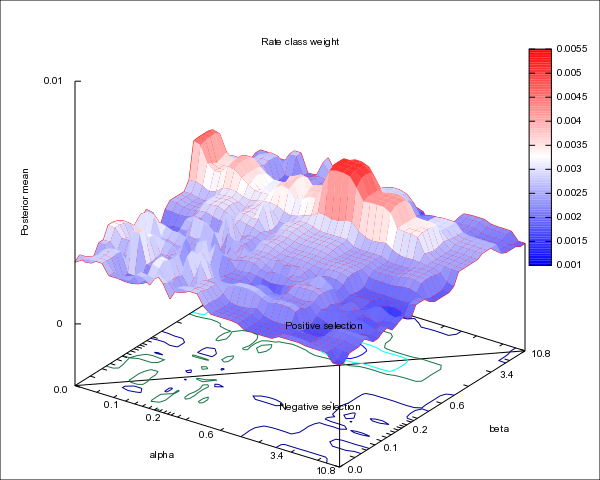
- This is a heat map showing sites are under positive selection and negative selection
PAML stuff
Identification of positive seleciton in the serine/threonine-protein kinase gene family
- In the evolution vertebrates, we would like to know if the branch leading to the Teleost fishes (genes A50 to A54)
You will need the following files
1. TF105351.Eut.3.phy
* this is the alignment file
2. TF105351.Eut.3.53876.tree * this is the newick tree with the branch of interest selected

3. TF105351.Eut.3.53876.ctl * CodeML configuration file for alternative model
4. TF105351.Eut.3.53876.fixed.ctl * CodeML configuration file for null model
Run the following commands
codeml TF105351.Eut.3.53876.ctl
codeml TF105351.Eut.3.53876.fixed.ctl
Analyze results
Get liklihood values
grep lnL TF105351.Eut.3.53876.mlc
lnL(ntime: 41 np: 46): -4707.209701 +0.000000
Liklihood value for alternative model is -4707.209701
grep lnL TF105351.Eut.3.53876.fixed.mlc
lnL(ntime: 41 np: 45): -4710.222252 +0.000000
Liklihood value for null model is -4710.222252
ΔLRT = 2×(lnL1 - lnL0) = 2×(-4707.209701 - (-4710.222252)) = 6.02578
The degree of freedom is 1 (np1 - np0 = 46 - 45). p-value = 0.01098 (under χ²) => significant.